Arsenic »
PDB 4cx1-4j8m »
4cx1 »
Arsenic in PDB 4cx1: Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine
Enzymatic activity of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine
All present enzymatic activity of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine:
1.14.13.39;
1.14.13.39;
Protein crystallography data
The structure of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine, PDB code: 4cx1
was solved by
H.Li,
T.L.Poulos,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 92.19 / 2.13 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.725, 106.199, 156.052, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.243 / 21.428 |
Other elements in 4cx1:
The structure of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine also contains other interesting chemical elements:
| Iron | (Fe) | 2 atoms |
| Zinc | (Zn) | 1 atom |
Arsenic Binding Sites:
The binding sites of Arsenic atom in the Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine
(pdb code 4cx1). This binding sites where shown within
5.0 Angstroms radius around Arsenic atom.
In total 2 binding sites of Arsenic where determined in the Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine, PDB code: 4cx1:
Jump to Arsenic binding site number: 1; 2;
In total 2 binding sites of Arsenic where determined in the Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine, PDB code: 4cx1:
Jump to Arsenic binding site number: 1; 2;
Arsenic binding site 1 out of 2 in 4cx1
Go back to
Arsenic binding site 1 out
of 2 in the Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine
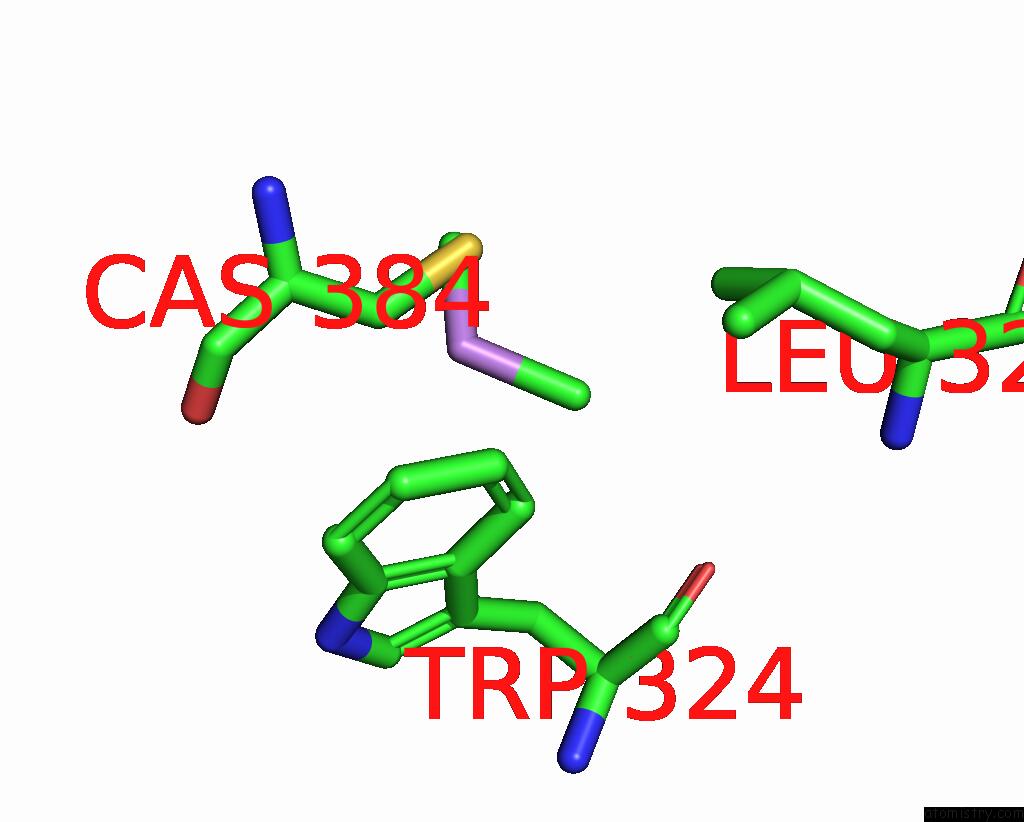
Mono view
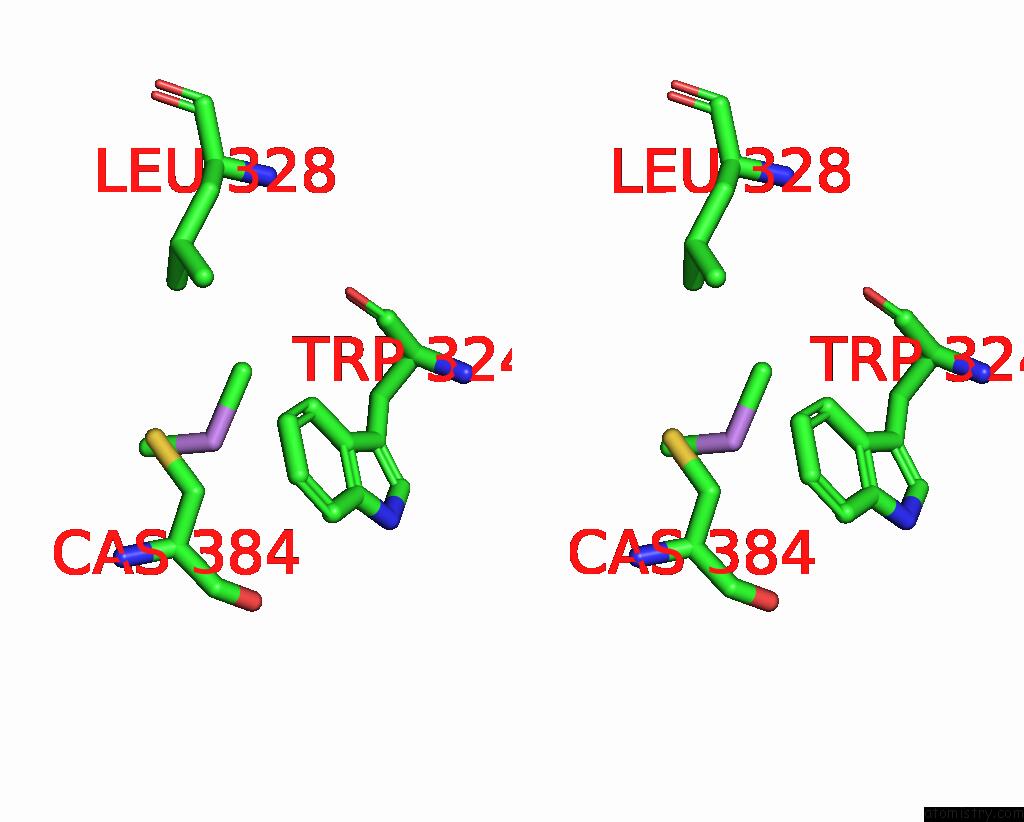
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 1 of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine within 5.0Å range:
|
Arsenic binding site 2 out of 2 in 4cx1
Go back to
Arsenic binding site 2 out
of 2 in the Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine
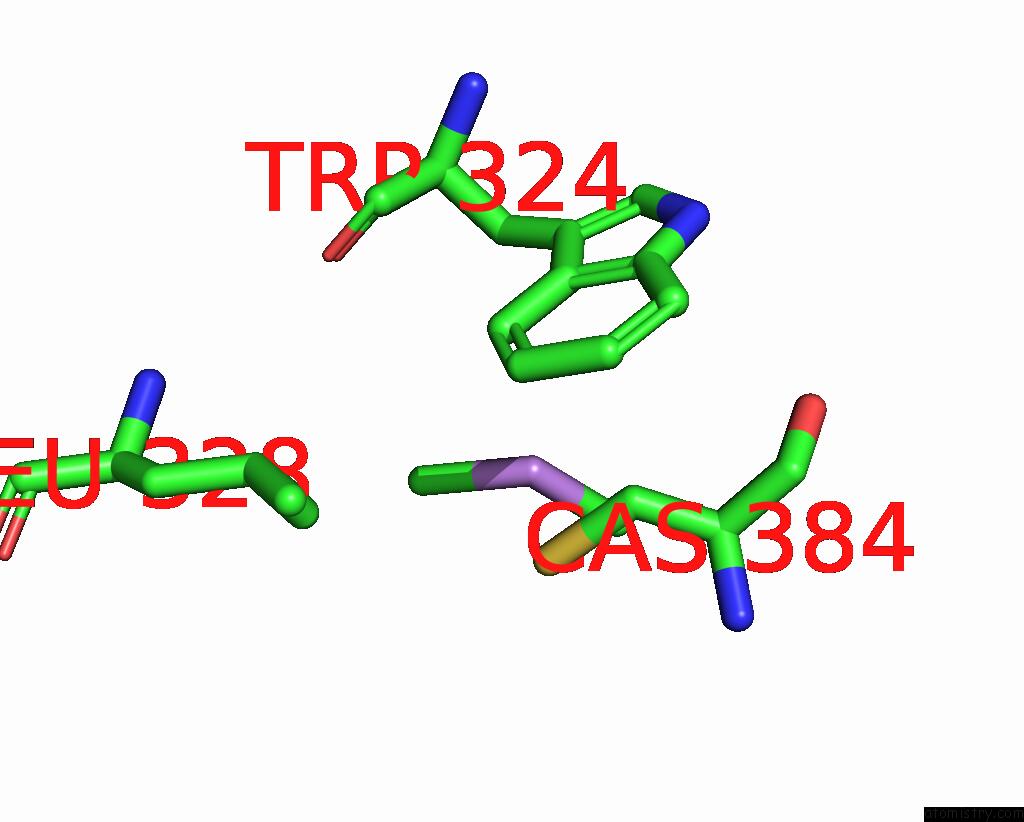
Mono view
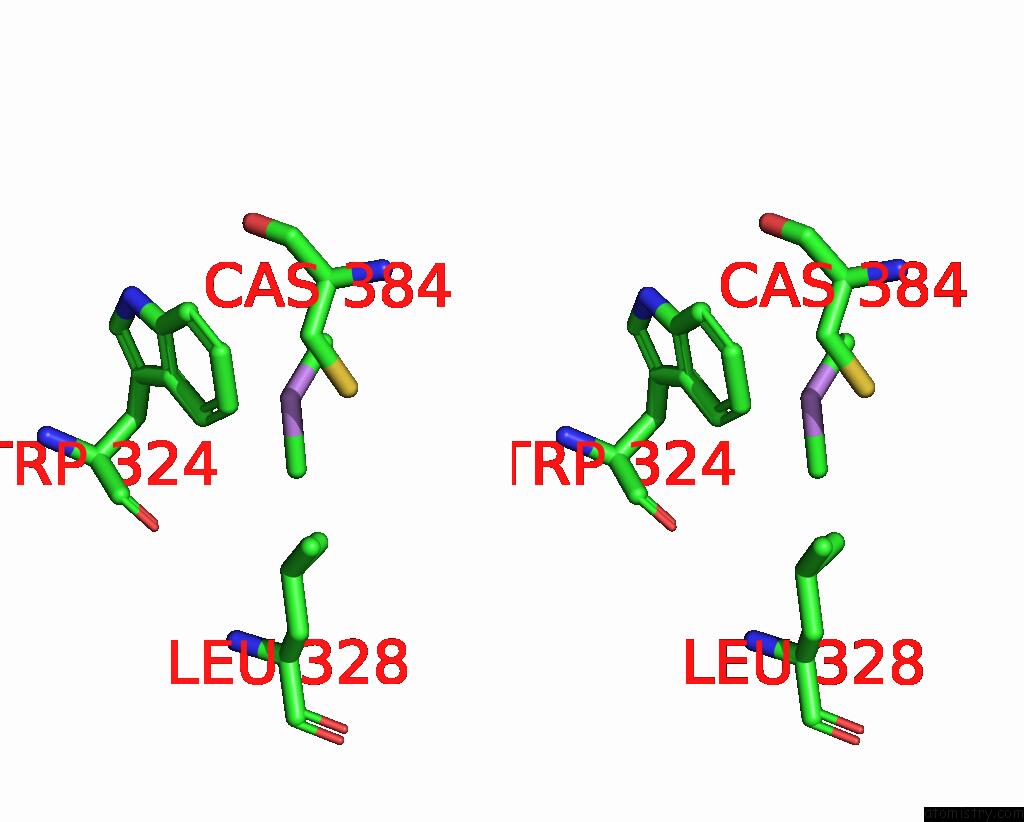
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 2 of Structure of Bovine Endothelial Nitric Oxide Synthase L111A Mutant Heme Domain in Complex with 4-Methyl-6-(((3R,4R)-4-( (5-(4-Methylpyridin-2-Yl)Pentyl)Oxy)Pyrrolidin-3-Yl)Methyl) Pyridin-2-Amine within 5.0Å range:
|
Reference:
H.Li,
J.Jamal,
S.L.Delker,
C.Plaza,
H.Ji,
Q.Jing,
H.Huang,
S.Kang,
R.B.Silverman,
T.L.Poulos.
Mobility of A Conserved Tyrosine Residue Controls Isoform- Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases. Biochemistry V. 53 5272 2014.
ISSN: ISSN 0006-2960
PubMed: 25089924
DOI: 10.1021/BI500561H
Page generated: Sun Jul 6 23:41:24 2025
ISSN: ISSN 0006-2960
PubMed: 25089924
DOI: 10.1021/BI500561H
Last articles
Cl in 2RCFCl in 2RC8
Cl in 2RBS
Cl in 2RB4
Cl in 2RBC
Cl in 2RAG
Cl in 2R8Y
Cl in 2RAS
Cl in 2RAY
Cl in 2RAB