Arsenic »
PDB 5hrn-5o8x »
5krt »
Arsenic in PDB 5krt: Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor
Protein crystallography data
The structure of Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor, PDB code: 5krt
was solved by
D.Patel,
J.D.Bauman,
E.Arnold,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.24 / 1.65 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.069, 72.069, 66.348, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.5 / 19.5 |
Other elements in 5krt:
The structure of Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor also contains other interesting chemical elements:
| Chlorine | (Cl) | 4 atoms |
Arsenic Binding Sites:
The binding sites of Arsenic atom in the Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor
(pdb code 5krt). This binding sites where shown within
5.0 Angstroms radius around Arsenic atom.
In total 2 binding sites of Arsenic where determined in the Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor, PDB code: 5krt:
Jump to Arsenic binding site number: 1; 2;
In total 2 binding sites of Arsenic where determined in the Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor, PDB code: 5krt:
Jump to Arsenic binding site number: 1; 2;
Arsenic binding site 1 out of 2 in 5krt
Go back to
Arsenic binding site 1 out
of 2 in the Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor
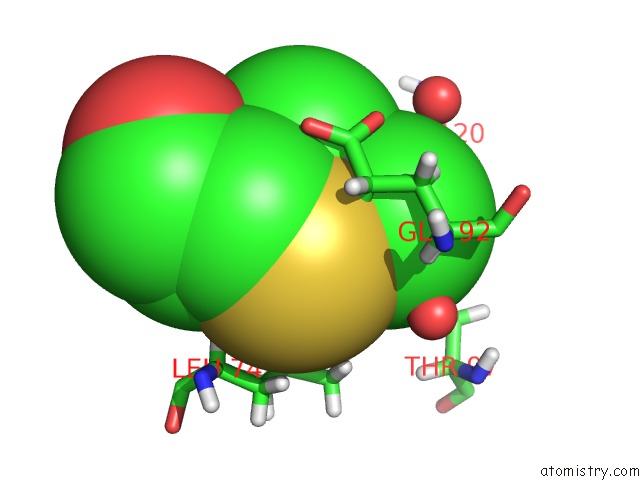
Mono view
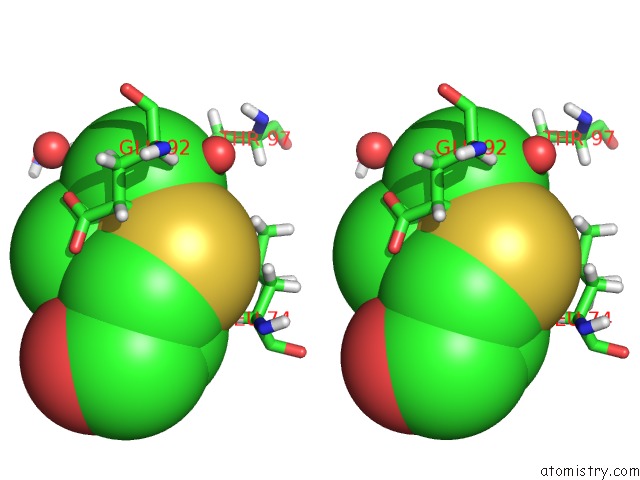
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 1 of Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor within 5.0Å range:
|
Arsenic binding site 2 out of 2 in 5krt
Go back to
Arsenic binding site 2 out
of 2 in the Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor
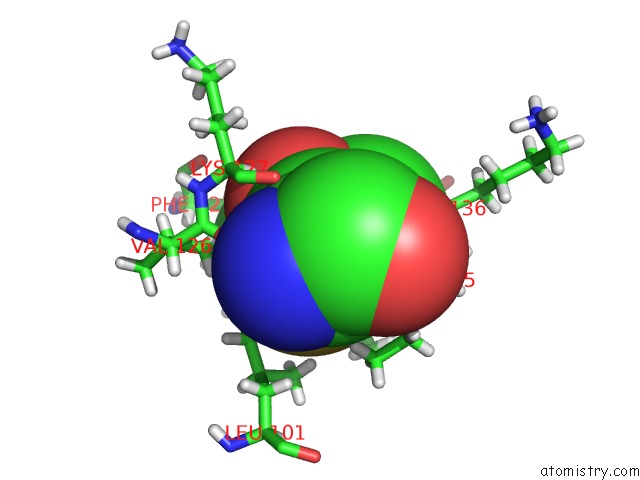
Mono view
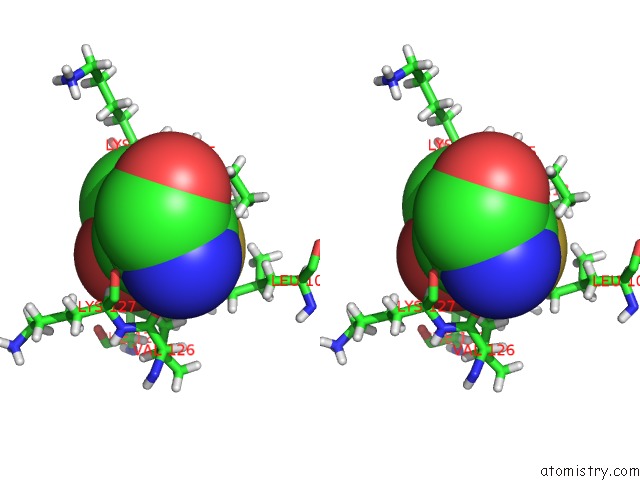
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 2 of Hiv-1 Integrase Catalytic Core Domain (Ccd) in Complex with A Fragment-Derived Allosteric Inhibitor within 5.0Å range:
|
Reference:
D.Patel,
J.Antwi,
P.C.Koneru,
E.Serrao,
S.Forli,
J.J.Kessl,
L.Feng,
N.Deng,
R.M.Levy,
J.R.Fuchs,
A.J.Olson,
A.N.Engelman,
J.D.Bauman,
M.Kvaratskhelia,
E.Arnold.
A New Class of Allosteric Hiv-1 Integrase Inhibitors Identified By Crystallographic Fragment Screening of the Catalytic Core Domain. J.Biol.Chem. V. 291 23569 2016.
ISSN: ESSN 1083-351X
PubMed: 27645997
DOI: 10.1074/JBC.M116.753384
Page generated: Mon Jul 7 00:18:27 2025
ISSN: ESSN 1083-351X
PubMed: 27645997
DOI: 10.1074/JBC.M116.753384
Last articles
Br in 3HOXBr in 3HOU
Br in 3HOW
Br in 3HOV
Br in 3HLL
Br in 3HNO
Br in 3H40
Br in 3H4B
Br in 3H4D
Br in 3GT3