Arsenic »
PDB 1glj-1pqu »
1pqu »
Arsenic in PDB 1pqu: Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate
Enzymatic activity of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate
All present enzymatic activity of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate:
1.2.1.11;
1.2.1.11;
Protein crystallography data
The structure of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate, PDB code: 1pqu
was solved by
J.Blanco,
R.A.Moore,
R.E.Viola,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.65 / 1.92 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.929, 76.579, 134.097, 90.00, 92.59, 90.00 |
| R / Rfree (%) | 18.2 / 23.4 |
Arsenic Binding Sites:
The binding sites of Arsenic atom in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate
(pdb code 1pqu). This binding sites where shown within
5.0 Angstroms radius around Arsenic atom.
In total 4 binding sites of Arsenic where determined in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate, PDB code: 1pqu:
Jump to Arsenic binding site number: 1; 2; 3; 4;
In total 4 binding sites of Arsenic where determined in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate, PDB code: 1pqu:
Jump to Arsenic binding site number: 1; 2; 3; 4;
Arsenic binding site 1 out of 4 in 1pqu
Go back to
Arsenic binding site 1 out
of 4 in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 1 of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate within 5.0Å range:
|
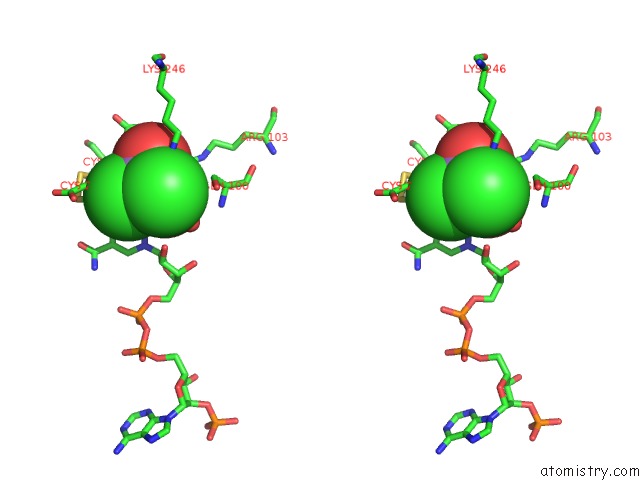
Arsenic binding site 2 out of 4 in 1pqu
Go back to
Arsenic binding site 2 out
of 4 in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 2 of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate within 5.0Å range:
|
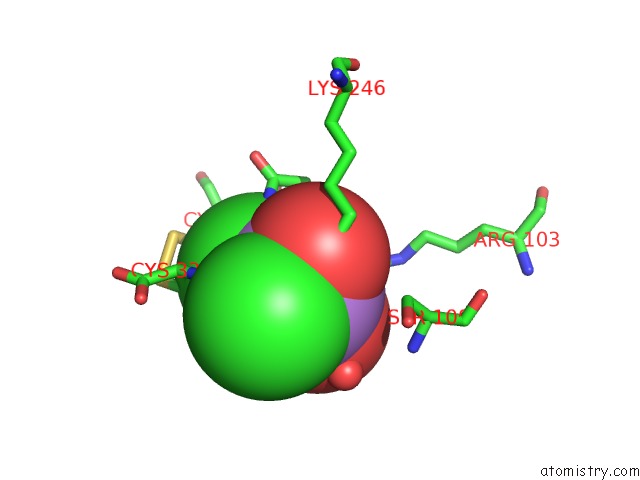
Arsenic binding site 3 out of 4 in 1pqu
Go back to
Arsenic binding site 3 out
of 4 in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 3 of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate within 5.0Å range:
|
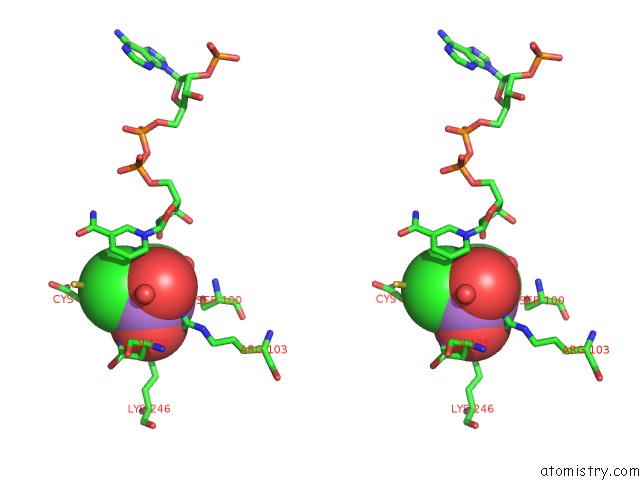
Arsenic binding site 4 out of 4 in 1pqu
Go back to
Arsenic binding site 4 out
of 4 in the Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Arsenic with other atoms in the As binding
site number 4 of Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase From Haemophilus Influenzae Bound with Nadp, S-Methyl Cysteine Sulfoxide and Cacodylate within 5.0Å range:
|
Reference:
J.Blanco,
R.A.Moore,
C.R.Faehnle,
D.M.Coe,
R.E.Viola.
The Role of Substrate-Binding Groups in the Mechanism of Aspartate-Beta-Semialdehyde Dehydrogenase. Acta Crystallogr.,Sect.D V. 60 1388 2004.
ISSN: ISSN 0907-4449
PubMed: 15272161
DOI: 10.1107/S0907444904012971
Page generated: Sun Jul 6 22:59:51 2025
ISSN: ISSN 0907-4449
PubMed: 15272161
DOI: 10.1107/S0907444904012971
Last articles
I in 4KICI in 4JK4
I in 4JXJ
I in 4JRX
I in 4JM9
I in 4J8F
I in 4J3U
I in 4JL9
I in 4J54
I in 4JIJ